Explore Fishing Data
ExploreFishingData.RmdOverview
A General overview of data. More detailed exploration based on Fleets, Ports, Species, can be found in other vignettes
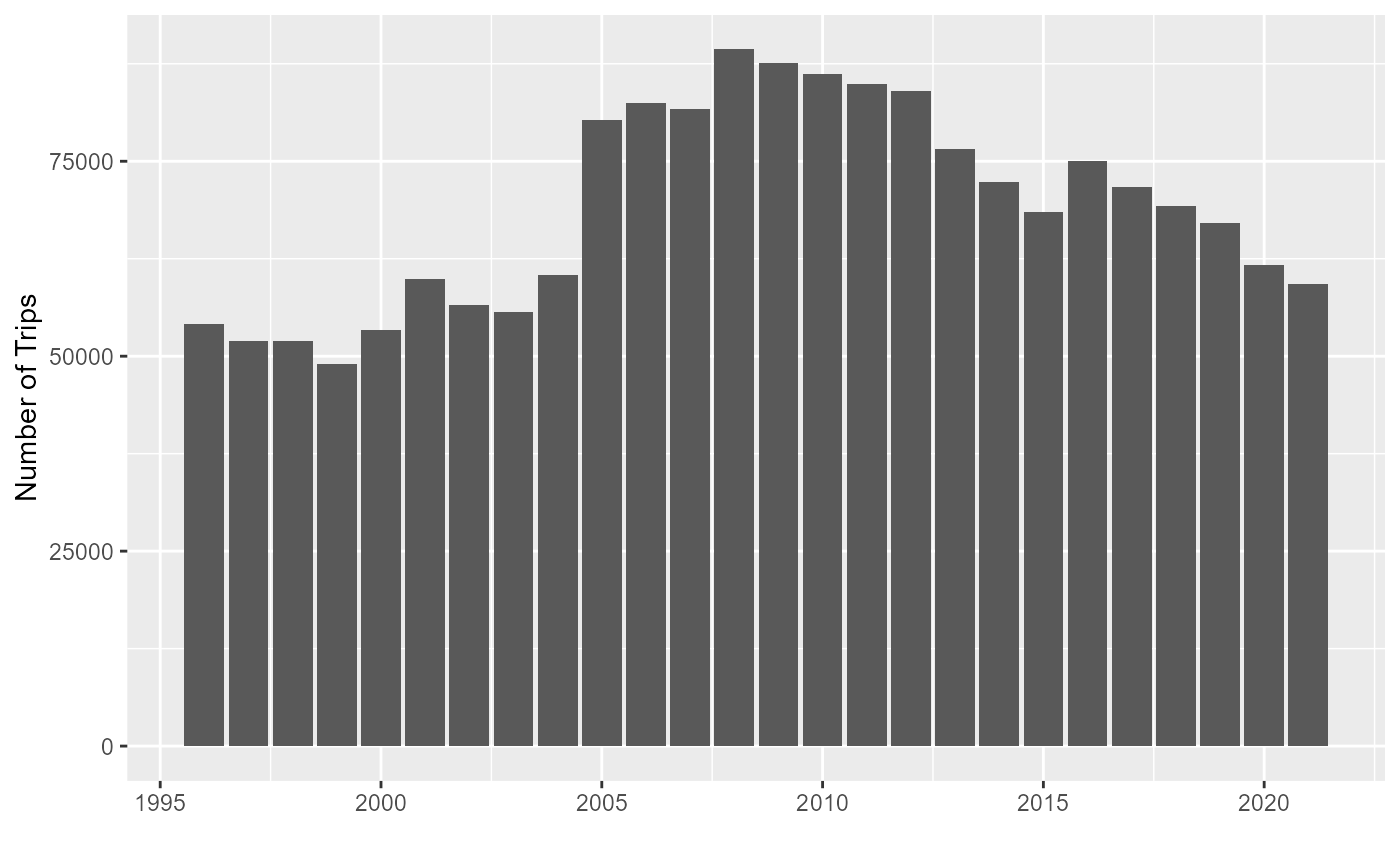
Data Sources and cleaning
Landings data by box at trip level provided by SSB for the communities at sea project
Example of data
Explain the fields
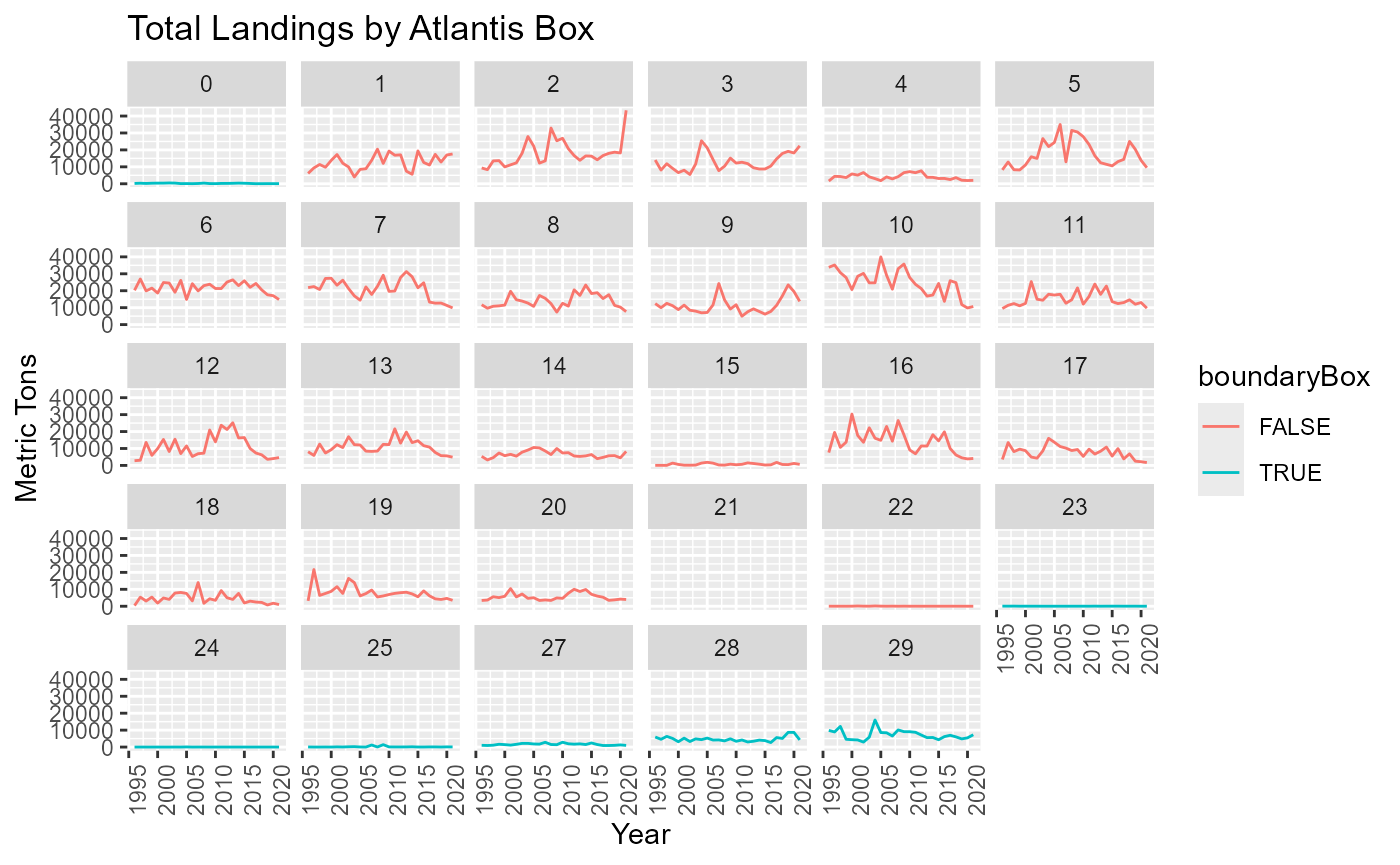
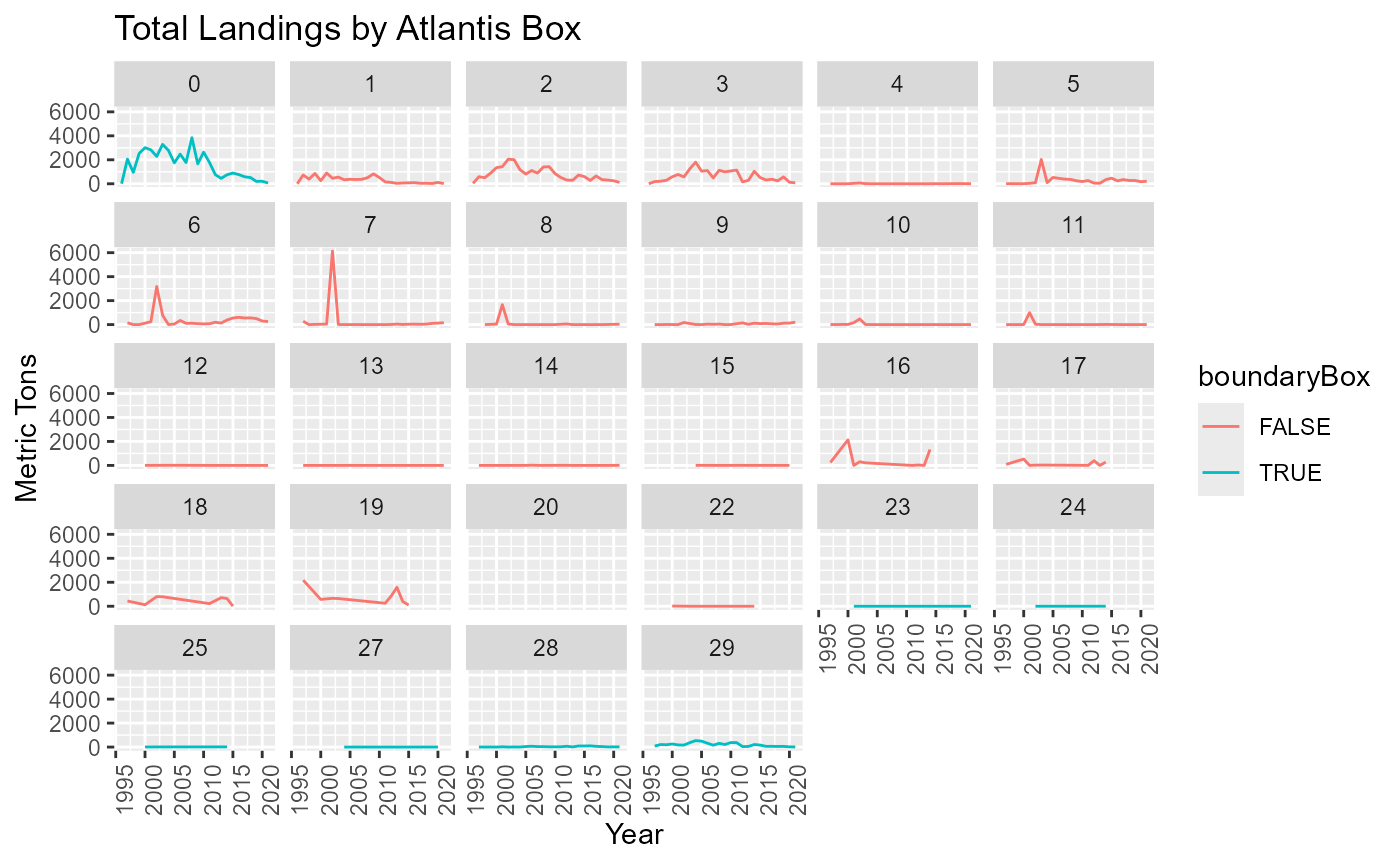
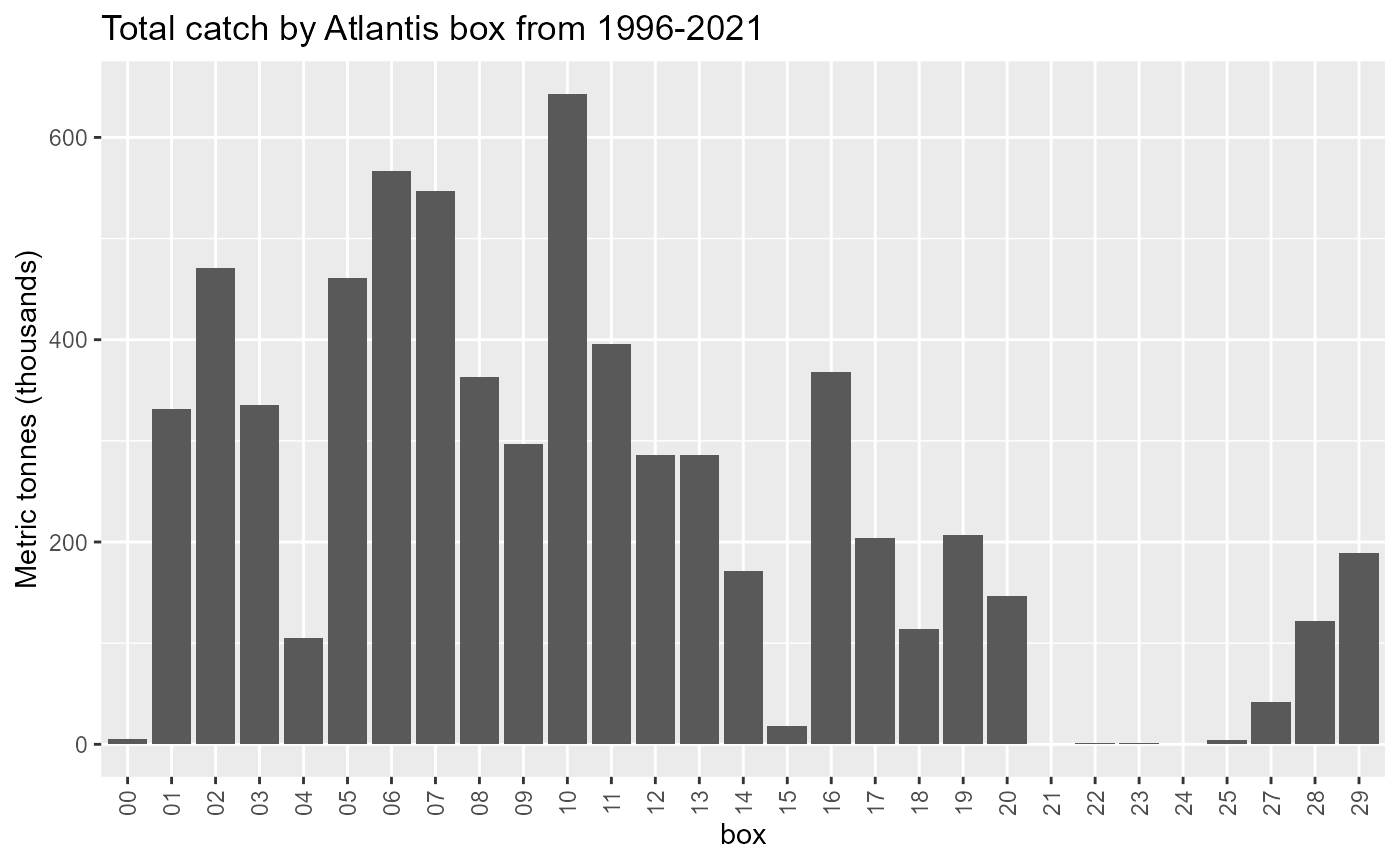
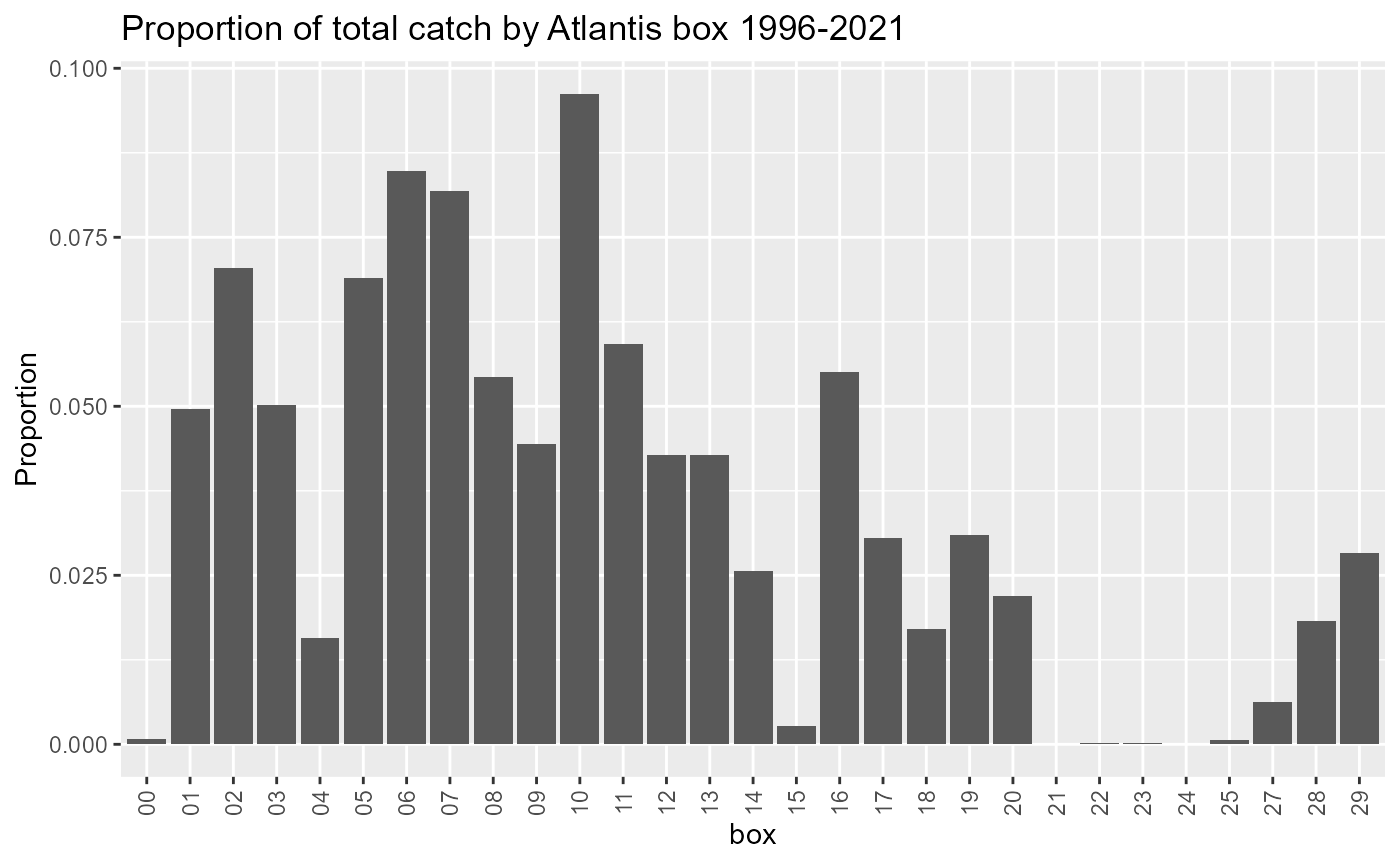
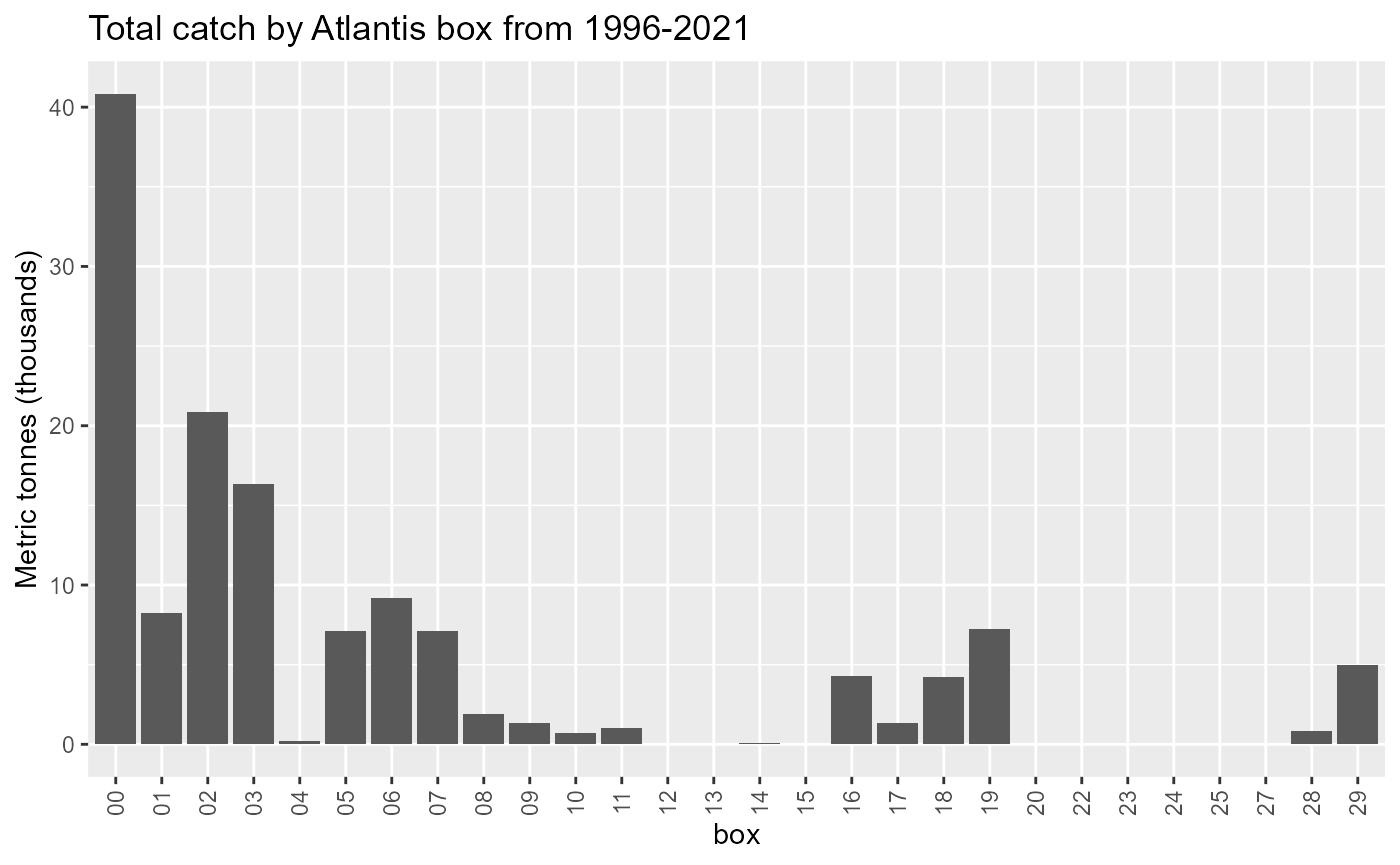
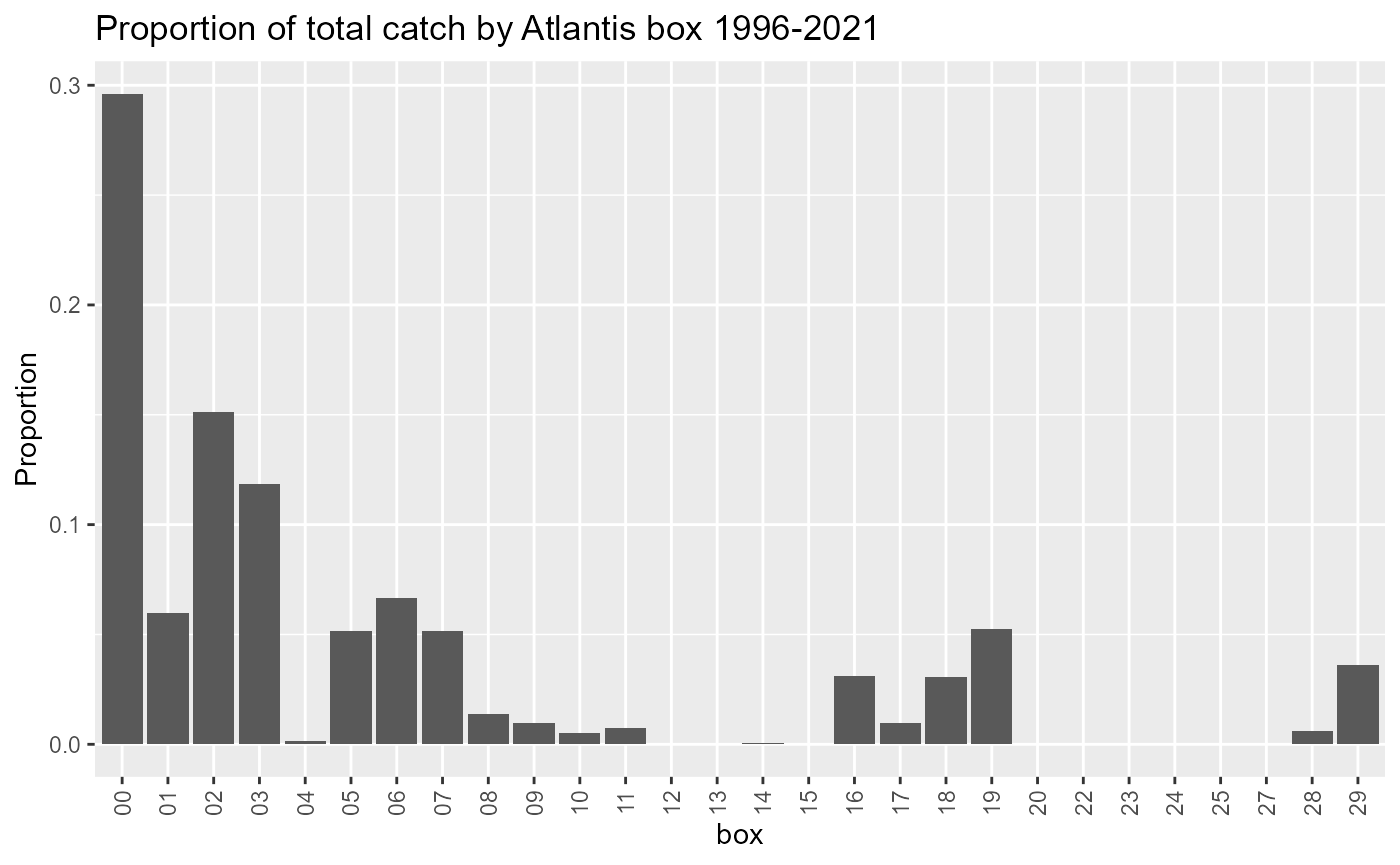
Landings in Boundary boxes
Atlantis boundary boxes are not modelled like other boxes so we need to know how much of the landings come from these boxes
Port level
There are 440 unique ports along the US eastern seaboard in which fish caught in the NEUS region are landed. 406 are within NEUS
The Main ports in terms of landings (1000’s metric tons). Only states within the model are considered
Landings by Ports and State
Only states within the model are considered Top 5 ports in each state based on landings (1000’s metric tons)
Ports by State
Lighly shaded bars indicate ports that lie outside of the NEUS footprint. Note: CN = Canadian ports
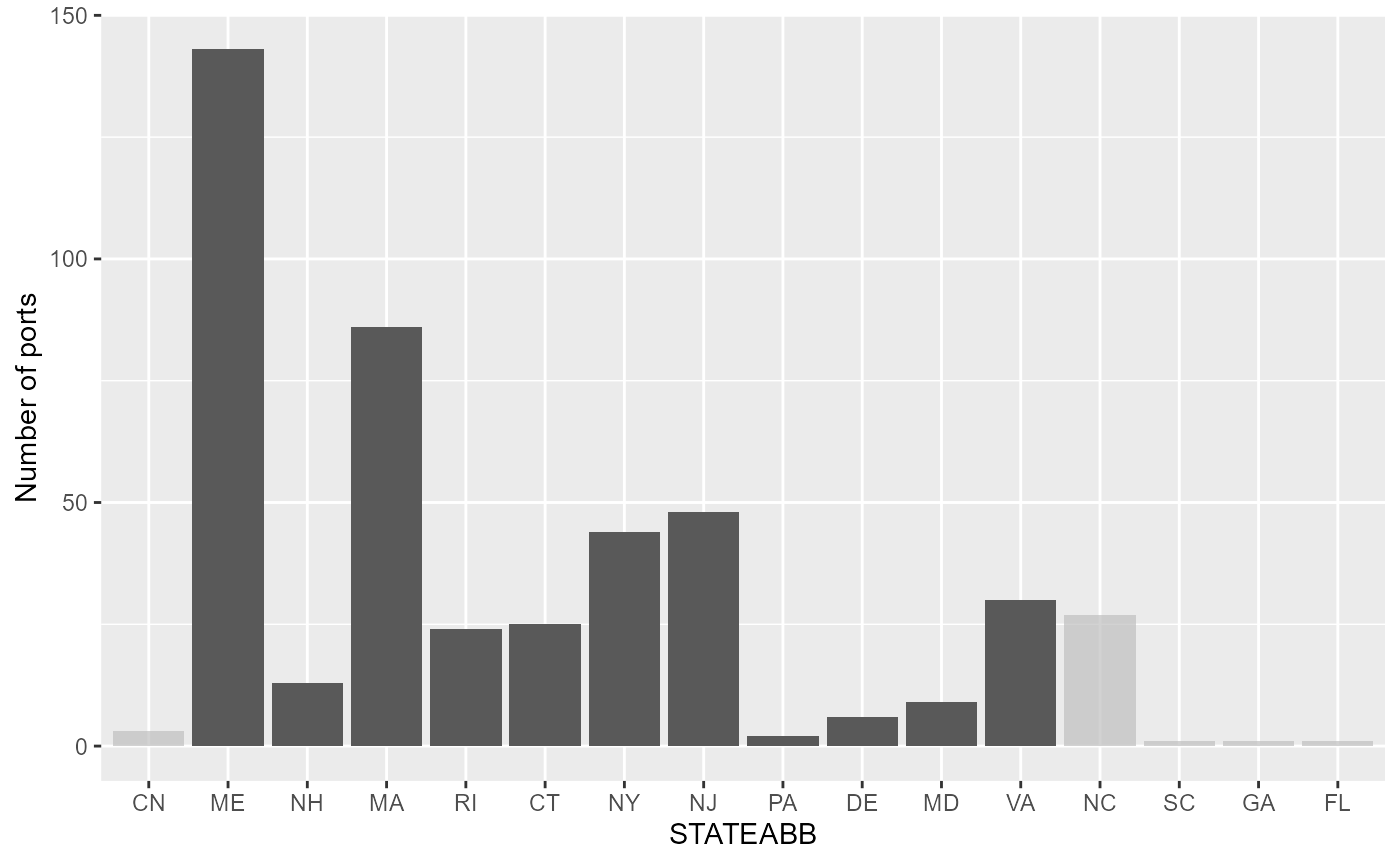
Are the fish landed from the boundary boxes distributed equally among ports or are some ports heavily reliant from landings in these boxes.
Landings by State
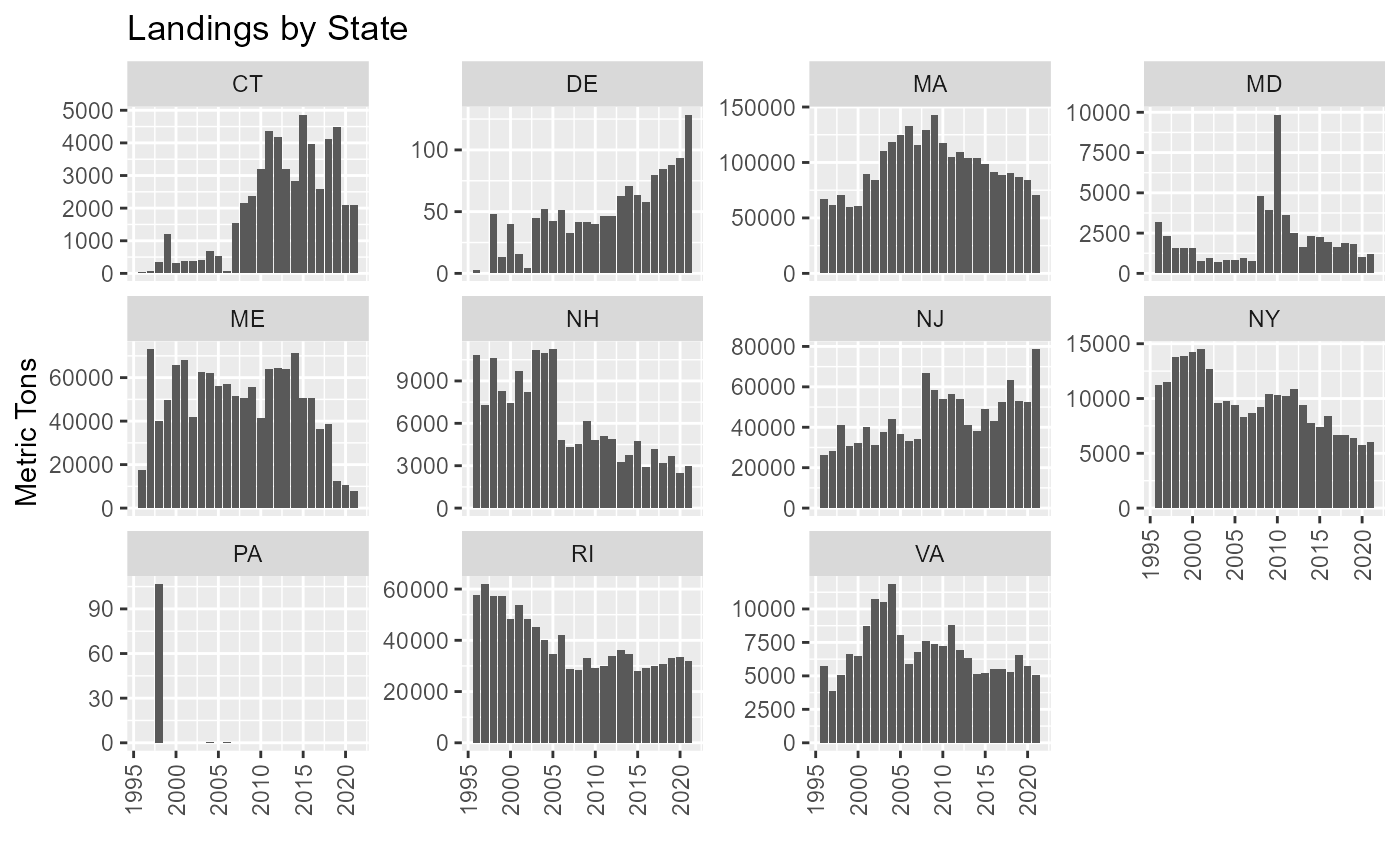
Missing State/port designations appear in data. This results in a number if trip not being able to be attributed to a port. Table shows number of trips with missing port/state information
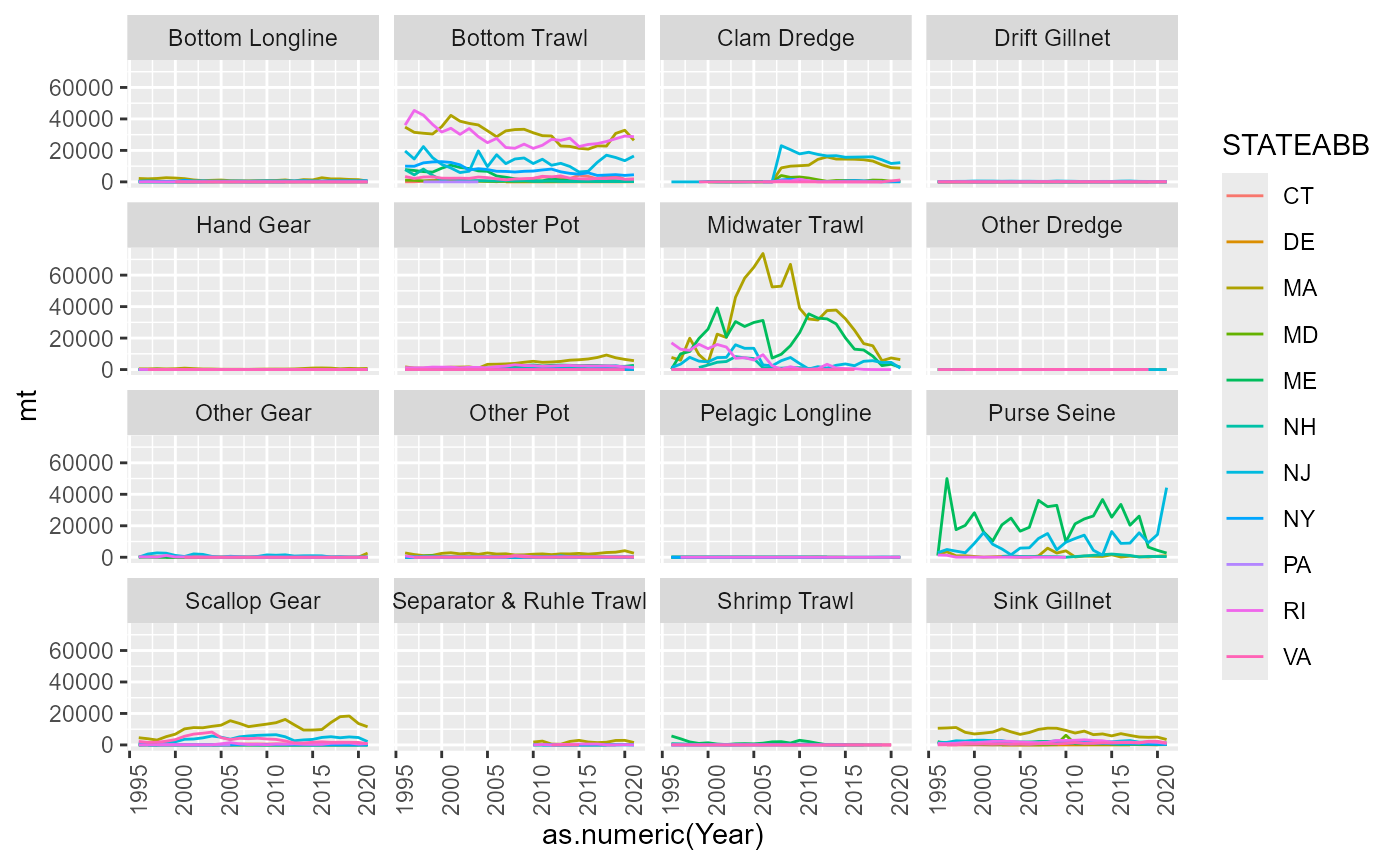
Gear categories
There are no GEARCODEs that have missing GEARCATs. The distinct Gear categories DMIS are :
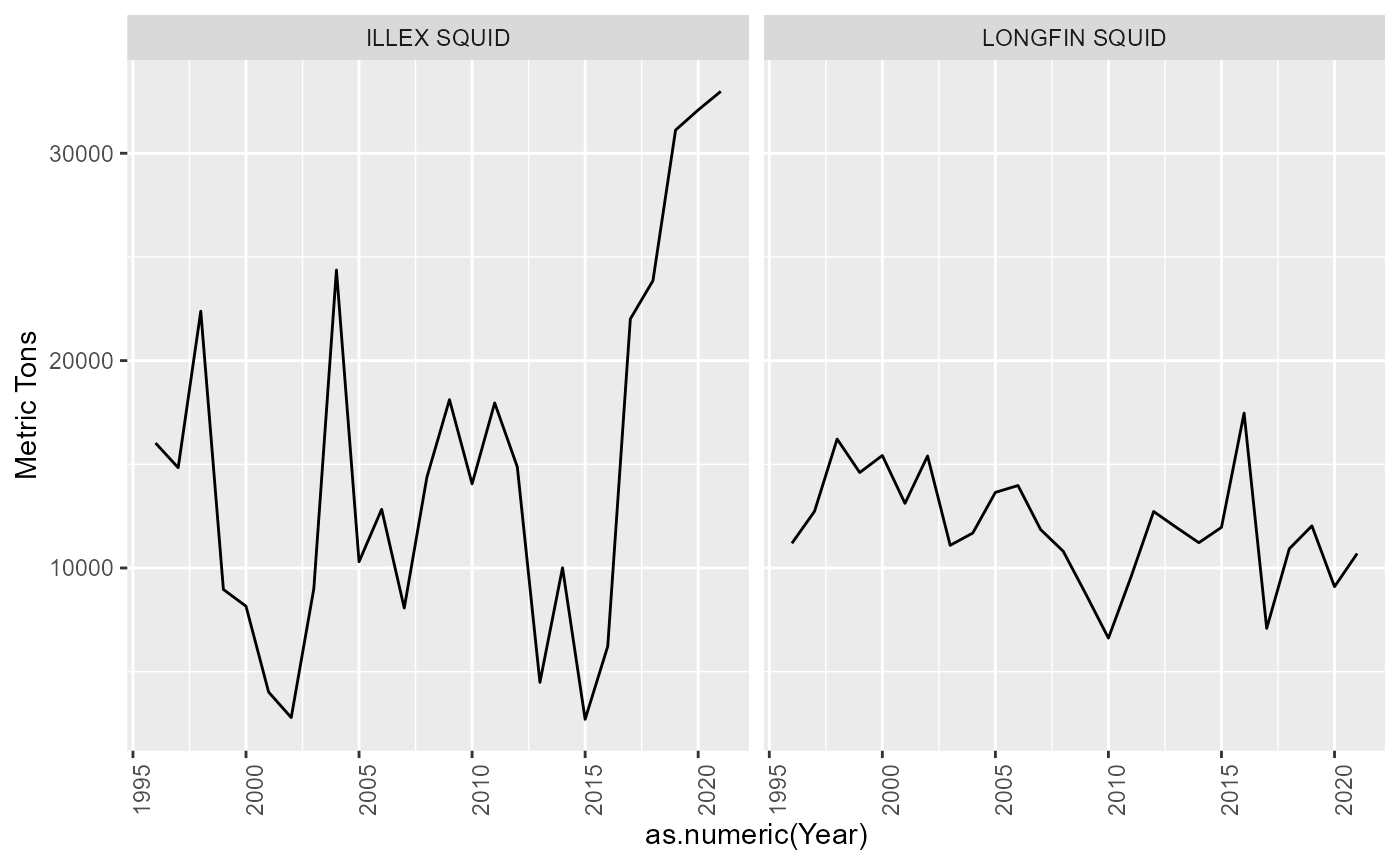
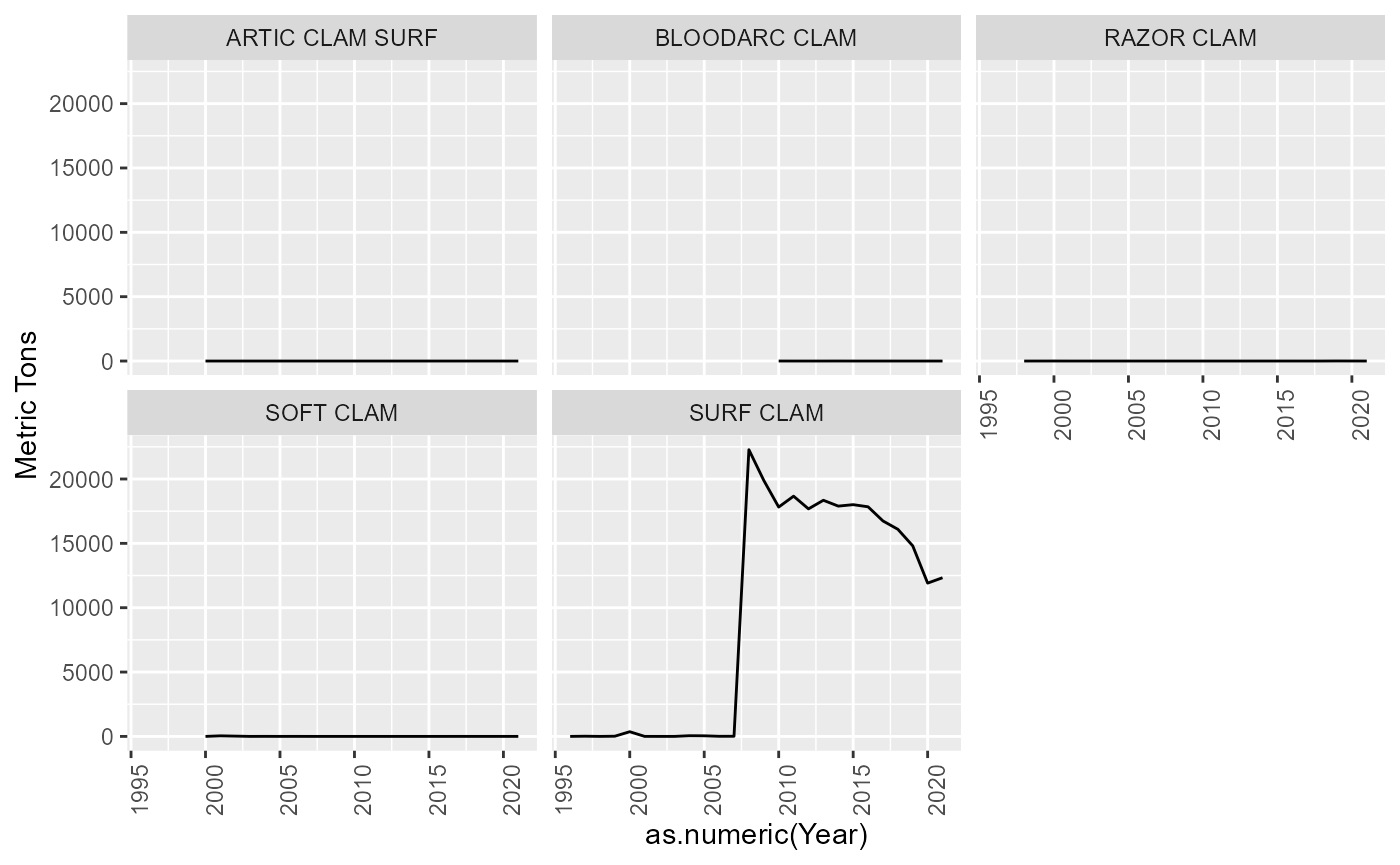
Species categories
There are some species without an atlantis code assigned. Some of these are “catch all” species codes that will need to be resolved. Eg. SKATES, NK SHARK, NK CLAM, OTHER FISH. Others can be ignored
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?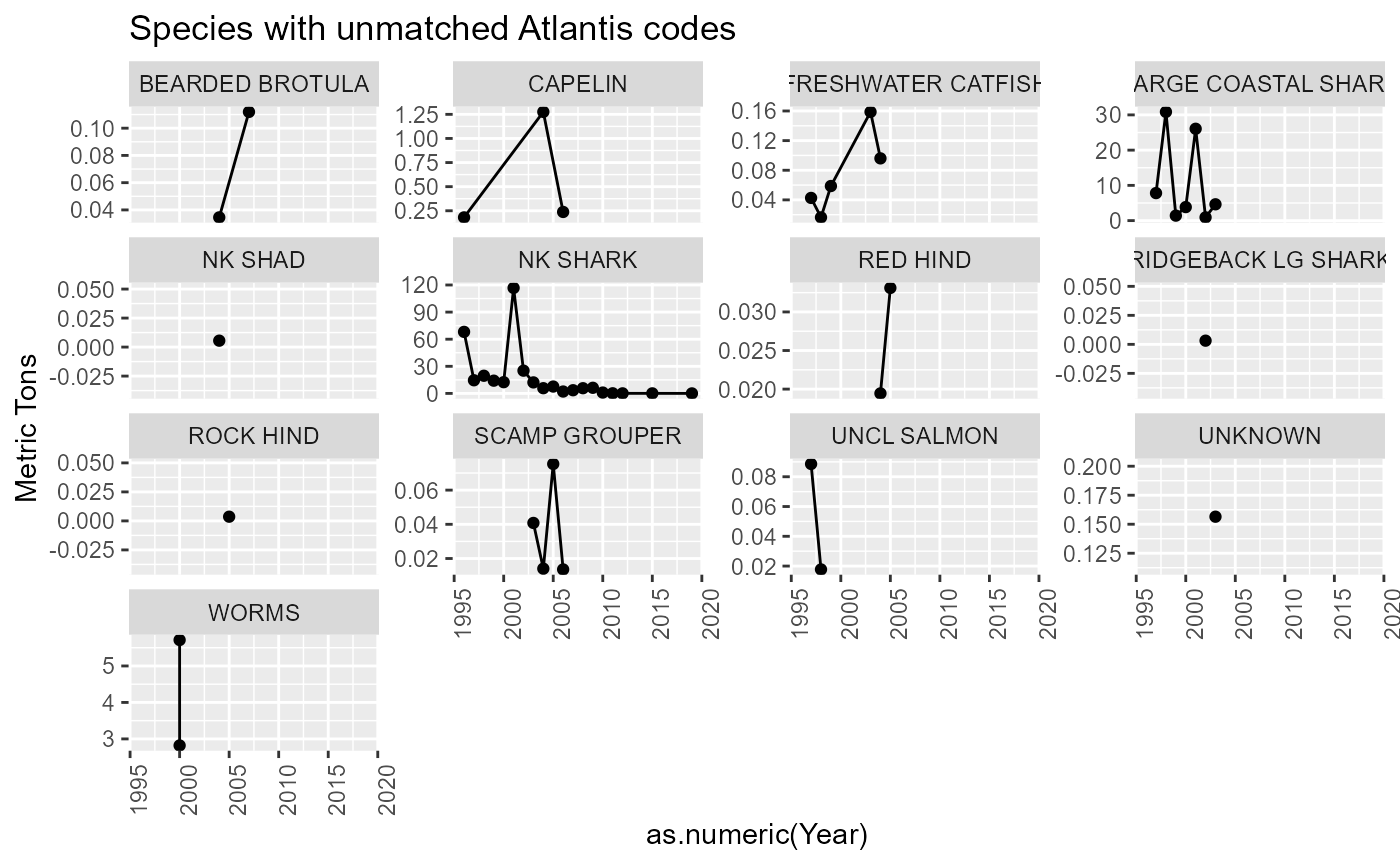
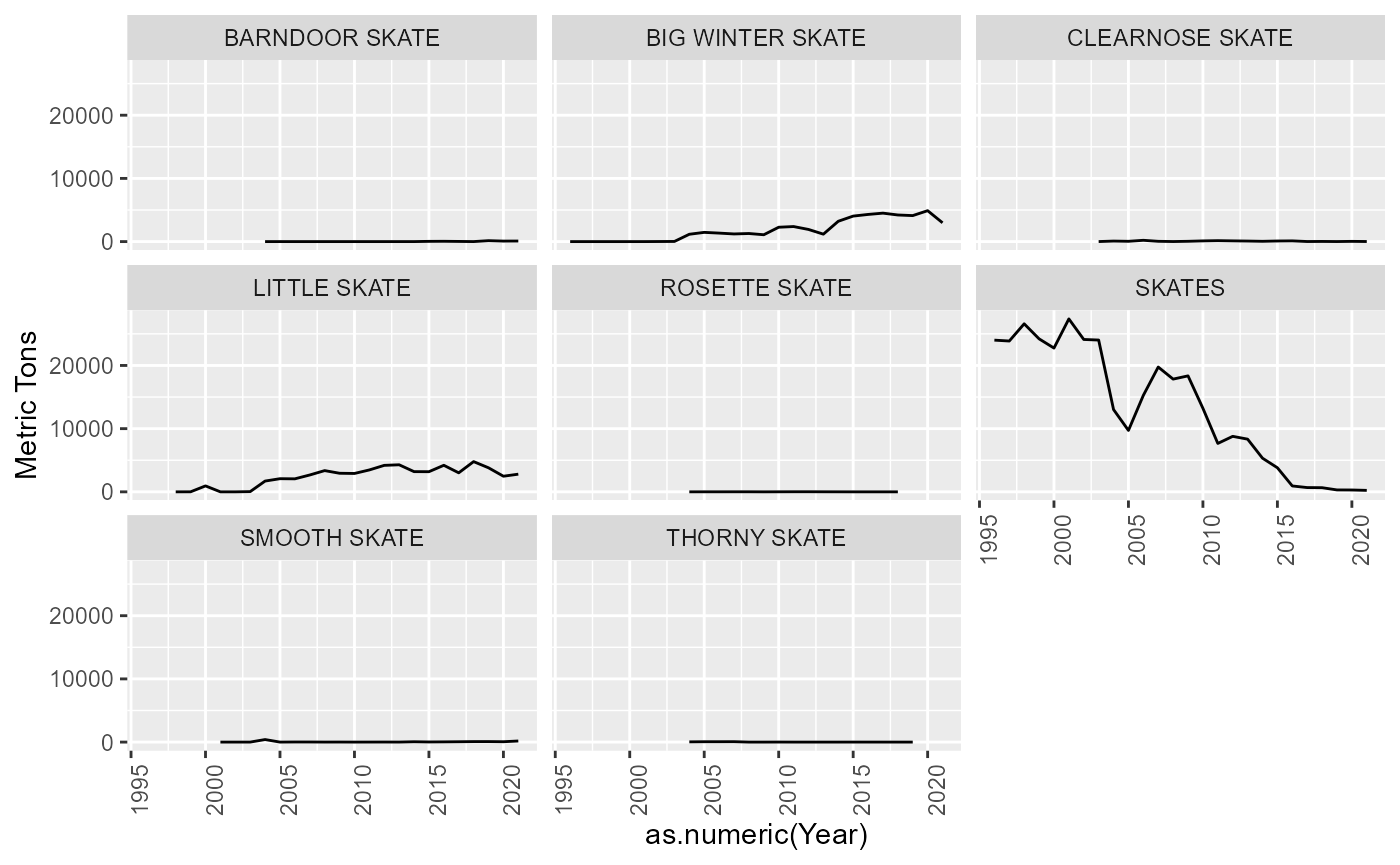
Skates
All species identified as a SKATE in the data. Unknown skates were
classed as “SKATES” and attributed to the Atlantis SK functional group
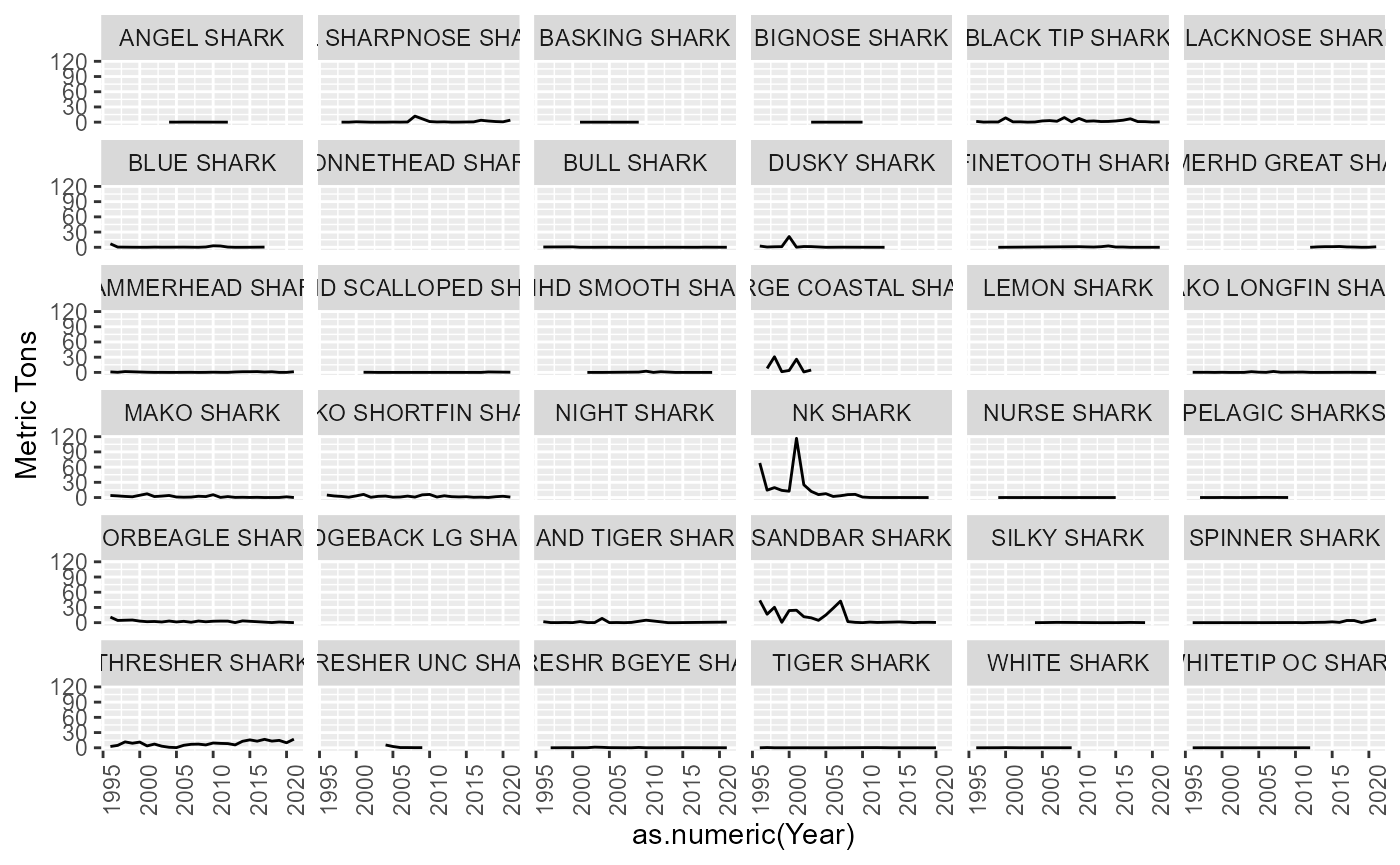
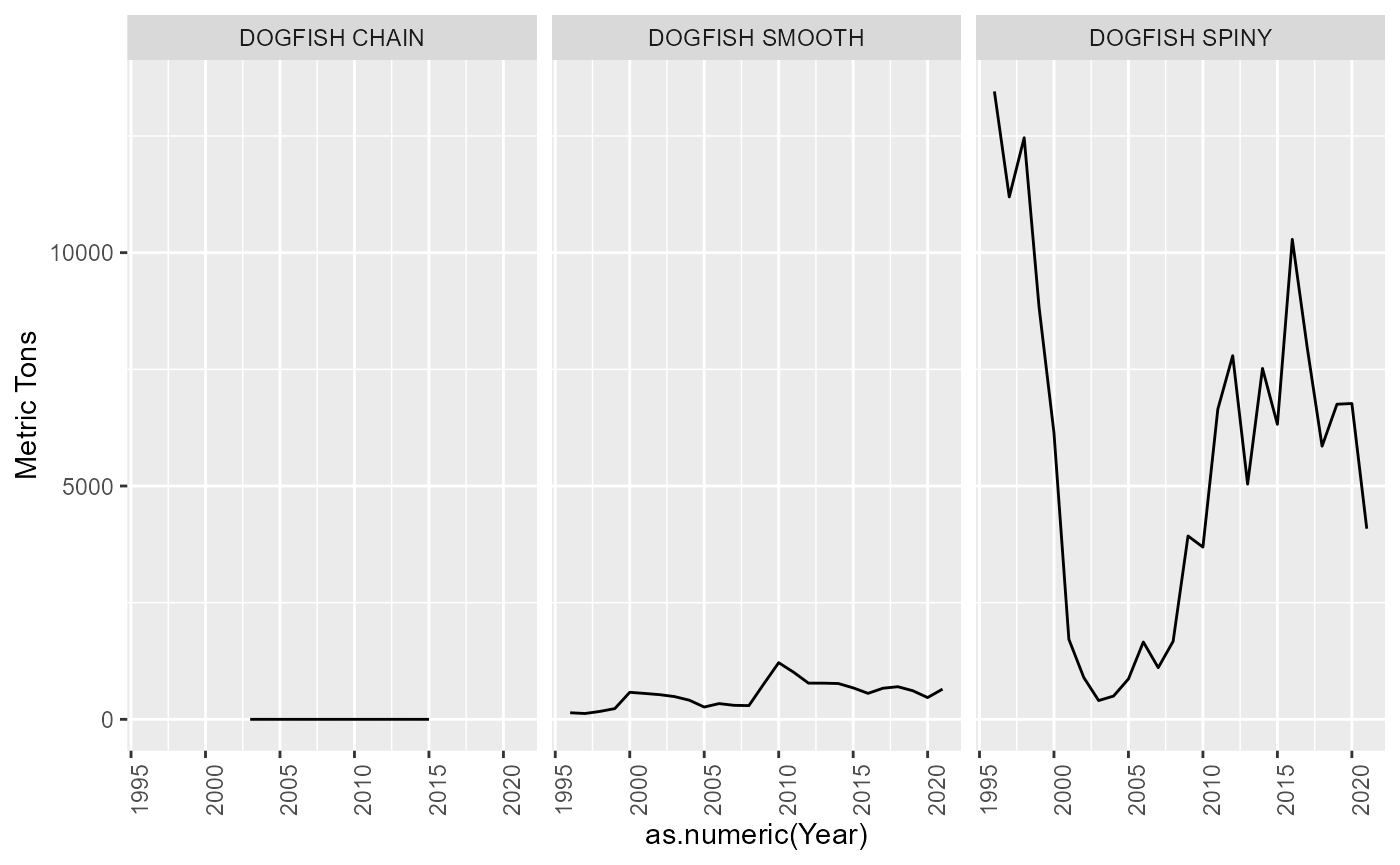
Shark
All species identified as a SHARK in the data. These were left as is
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?
#> `geom_line()`: Each group consists of only one observation.
#> ℹ Do you need to adjust the group aesthetic?