Setting up a food web model
First, install Rpath as instructed in Getting Started. R package data.table is also required to run the code in this article.
Parameter file generation
Unlike the GUI based EwE software package, Rpath relies on a
parameter input file. This file is actually a list of several different
parameter files: model, diet, stanzas, and pedigree. Parameter files can
be created outside of R and read in using the
read.rpath.params() function. This function will merge
several different flat files into an R object of the list type. An
alternative is to generate the list file and populate it completely
within R. The function create.rpath.params() will generate
an Rpath.params list object. This ensures that all of the
correct columns are present in the parameter file.
Additionally, an existing model can be imported from EwE, by first
exporting the model from EwE in .eiixml format via the GUI interface,
and using the create.rpath.from.eiixml function to read in
the exported file (see the vignette
Convert_EwE_to_Rpath).
The parameter file contains all of the information you would normally enter in the input data tabs in EwE. There are 2 necessary pieces of information to generate the parameter file: the group names and their corresponding type. The types are: living = 0, primary producer = 1, detritus = 2, and fleet = 3. If your model contains multi-stanza groups then you need 2 additional pieces of information: stanza group names (include NA for those groups not in a stanza) and the number of stanzas per stanza group.
# Groups and types for the R Ecosystem
groups <- c('Seabirds', 'Whales', 'Seals', 'JuvRoundfish1', 'AduRoundfish1',
'JuvRoundfish2', 'AduRoundfish2', 'JuvFlatfish1', 'AduFlatfish1',
'JuvFlatfish2', 'AduFlatfish2', 'OtherGroundfish', 'Foragefish1',
'Foragefish2', 'OtherForagefish', 'Megabenthos', 'Shellfish',
'Macrobenthos', 'Zooplankton', 'Phytoplankton', 'Detritus',
'Discards', 'Trawlers', 'Midwater', 'Dredgers')
types <- c(rep(0, 19), 1, rep(2, 2), rep(3, 3))
stgroups <- c(rep(NA, 3), rep('Roundfish1', 2), rep('Roundfish2', 2),
rep('Flatfish1', 2), rep('Flatfish2', 2), rep(NA, 14))
REco.params <- create.rpath.params(group = groups, type = types, stgroup = stgroups)REco.params now contains a list of 4 objects: model,
diet, stanzas, and pedigree. The majority of the parameters are
populated with NA save those that have logical default vaules (i.e.,
0.66667 for VBGF_d).
Model parameters
The model parameter list contains the biomass, production to biomass, consumption to biomass, etc. parameters as well as the detrital fate parameters and fleet landings and discards.
| Group | Type | Biomass | PB | QB | EE | ProdCons | BioAcc | Unassim | DetInput | Detritus | Discards | Trawlers | Midwater | Dredgers | Trawlers.disc | Midwater.disc | Dredgers.disc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Seabirds | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| Whales | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| Seals | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| JuvRoundfish1 | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| AduRoundfish1 | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| JuvRoundfish2 | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| AduRoundfish2 | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| JuvFlatfish1 | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| AduFlatfish1 | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| JuvFlatfish2 | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| AduFlatfish2 | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| OtherGroundfish | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| Foragefish1 | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| Foragefish2 | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| OtherForagefish | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| Megabenthos | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| Shellfish | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| Macrobenthos | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| Zooplankton | 0 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| Phytoplankton | 1 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| Detritus | 2 | NA | NA | NA | NA | NA | NA | NA | 0 | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| Discards | 2 | NA | NA | NA | NA | NA | NA | NA | 0 | NA | NA | 0 | 0 | 0 | 0 | 0 | 0 |
| Trawlers | 3 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Midwater | 3 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Dredgers | 3 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
Each of the parameter lists are data tables (with the exception of the stanzas list which is itself a list of an integer and two data tables). Data tables are an extension of the classic data frame class. Advantages of data tables include simplified indexing which eases the process of populating the parameters. For example, you can add data to a specific slot or fill an entire column.
# Example of filling specific slots
REco.params$model[Group %in% c('Seals', 'Megabenthos'), EE := 0.8]
# Example of filling an entire column
biomass <- c(0.0149, 0.454, NA, NA, 1.39, NA, 5.553, NA, 5.766, NA,
0.739, 7.4, 5.1, 4.7, 5.1, NA, 7, 17.4, 23, 10, rep(NA, 5))
REco.params$model[, Biomass := biomass]Note the use of the operator ‘:=’ to assign values. This is unique to data tables from data.table.
| Group | Type | Biomass | EE |
|---|---|---|---|
| Seabirds | 0 | 0.0149 | NA |
| Whales | 0 | 0.4540 | NA |
| Seals | 0 | NA | 0.8 |
| JuvRoundfish1 | 0 | NA | NA |
| AduRoundfish1 | 0 | 1.3900 | NA |
| JuvRoundfish2 | 0 | NA | NA |
| AduRoundfish2 | 0 | 5.5530 | NA |
| JuvFlatfish1 | 0 | NA | NA |
| AduFlatfish1 | 0 | 5.7660 | NA |
| JuvFlatfish2 | 0 | NA | NA |
| AduFlatfish2 | 0 | 0.7390 | NA |
| OtherGroundfish | 0 | 7.4000 | NA |
| Foragefish1 | 0 | 5.1000 | NA |
| Foragefish2 | 0 | 4.7000 | NA |
| OtherForagefish | 0 | 5.1000 | NA |
| Megabenthos | 0 | NA | 0.8 |
| Shellfish | 0 | 7.0000 | NA |
| Macrobenthos | 0 | 17.4000 | NA |
| Zooplankton | 0 | 23.0000 | NA |
| Phytoplankton | 1 | 10.0000 | NA |
| Detritus | 2 | NA | NA |
| Discards | 2 | NA | NA |
| Trawlers | 3 | NA | NA |
| Midwater | 3 | NA | NA |
| Dredgers | 3 | NA | NA |
Here are the rest of the columns for the model list.
# Model
biomass <- c(0.0149, 0.454, NA, NA, 1.39, NA, 5.553, NA, 5.766, NA,
0.739, 7.4, 5.1, 4.7, 5.1, NA, 7, 17.4, 23, 10, rep(NA, 5))
pb <- c(0.098, 0.031, 0.100, 2.026, 0.42, 2.1, 0.425, 1.5, 0.26, 1.1, 0.18, 0.6,
0.61, 0.65, 1.5, 0.9, 1.3, 7, 39, 240, rep(NA, 5))
qb <- c(76.750, 6.976, 34.455, NA, 2.19, NA, 3.78, NA, 1.44, NA, 1.69,
1.764, 3.52, 5.65, 3.6, 2.984, rep (NA, 9))
REco.params$model[, Biomass := biomass]
REco.params$model[, PB := pb]
REco.params$model[, QB := qb]
# EE for groups without biomass
REco.params$model[Group %in% c('Seals', 'Megabenthos'), EE := 0.8]
# Production to Consumption for those groups without a QB
REco.params$model[Group %in% c('Shellfish', 'Zooplankton'), ProdCons:= 0.25]
REco.params$model[Group == 'Macrobenthos', ProdCons := 0.35]
# Biomass accumulation and unassimilated consumption
REco.params$model[, BioAcc := c(rep(0, 22), rep(NA, 3))]
REco.params$model[, Unassim := c(rep(0.2, 18), 0.4, rep(0, 3), rep(NA, 3))]
# Detrital Fate
REco.params$model[, Detritus := c(rep(1, 20), rep(0, 5))]
REco.params$model[, Discards := c(rep(0, 22), rep(1, 3))]
# Fisheries
# Landings
trawl <- c(rep(0, 4), 0.08, 0, 0.32, 0, 0.09, 0, 0.05, 0.2, rep(0, 10), rep(NA, 3))
mid <- c(rep(0, 12), 0.3, 0.08, 0.02, rep(0, 7), rep(NA, 3))
dredge <- c(rep(0, 15), 0.1, 0.5, rep(0, 5), rep(NA, 3))
REco.params$model[, Trawlers := trawl]
REco.params$model[, Midwater := mid]
REco.params$model[, Dredgers := dredge]
# Discards
trawl.d <- c(1e-5, 1e-7, 0.001, 0.001, 0.005, 0.001, 0.009, 0.001, 0.04, 0.001,
0.01, 0.08, 0.001, 0.001, 0.001, rep(0, 7), rep(NA, 3))
mid.d <- c(rep(0, 2), 0.001, 0.001, 0.01, 0.001, 0.01, rep(0, 4), 0.05, 0.05,
0.01, 0.01, rep(0, 7), rep(NA, 3))
dredge.d <- c(rep(0, 3), 0.001, 0.05, 0.001, 0.05, 0.001, 0.05, 0.001, 0.01, 0.05,
rep(0, 3), 0.09, 0.01, 1e-4, rep(0, 4), rep(NA, 3))
REco.params$model[, Trawlers.disc := trawl.d]
REco.params$model[, Midwater.disc := mid.d]
REco.params$model[, Dredgers.disc := dredge.d]| Group | Type | Biomass | PB | QB | EE | ProdCons | BioAcc | Unassim | DetInput | Detritus | Discards | Trawlers | Midwater | Dredgers | Trawlers.disc | Midwater.disc | Dredgers.disc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Seabirds | 0 | 0.0149 | 0.098 | 76.750 | NA | NA | 0 | 0.2 | NA | 1 | 0 | 0.00 | 0.00 | 0.0 | 1e-05 | 0.000 | 0e+00 |
| Whales | 0 | 0.4540 | 0.031 | 6.976 | NA | NA | 0 | 0.2 | NA | 1 | 0 | 0.00 | 0.00 | 0.0 | 1e-07 | 0.000 | 0e+00 |
| Seals | 0 | NA | 0.100 | 34.455 | 0.8 | NA | 0 | 0.2 | NA | 1 | 0 | 0.00 | 0.00 | 0.0 | 1e-03 | 0.001 | 0e+00 |
| JuvRoundfish1 | 0 | NA | 2.026 | NA | NA | NA | 0 | 0.2 | NA | 1 | 0 | 0.00 | 0.00 | 0.0 | 1e-03 | 0.001 | 1e-03 |
| AduRoundfish1 | 0 | 1.3900 | 0.420 | 2.190 | NA | NA | 0 | 0.2 | NA | 1 | 0 | 0.08 | 0.00 | 0.0 | 5e-03 | 0.010 | 5e-02 |
| JuvRoundfish2 | 0 | NA | 2.100 | NA | NA | NA | 0 | 0.2 | NA | 1 | 0 | 0.00 | 0.00 | 0.0 | 1e-03 | 0.001 | 1e-03 |
| AduRoundfish2 | 0 | 5.5530 | 0.425 | 3.780 | NA | NA | 0 | 0.2 | NA | 1 | 0 | 0.32 | 0.00 | 0.0 | 9e-03 | 0.010 | 5e-02 |
| JuvFlatfish1 | 0 | NA | 1.500 | NA | NA | NA | 0 | 0.2 | NA | 1 | 0 | 0.00 | 0.00 | 0.0 | 1e-03 | 0.000 | 1e-03 |
| AduFlatfish1 | 0 | 5.7660 | 0.260 | 1.440 | NA | NA | 0 | 0.2 | NA | 1 | 0 | 0.09 | 0.00 | 0.0 | 4e-02 | 0.000 | 5e-02 |
| JuvFlatfish2 | 0 | NA | 1.100 | NA | NA | NA | 0 | 0.2 | NA | 1 | 0 | 0.00 | 0.00 | 0.0 | 1e-03 | 0.000 | 1e-03 |
| AduFlatfish2 | 0 | 0.7390 | 0.180 | 1.690 | NA | NA | 0 | 0.2 | NA | 1 | 0 | 0.05 | 0.00 | 0.0 | 1e-02 | 0.000 | 1e-02 |
| OtherGroundfish | 0 | 7.4000 | 0.600 | 1.764 | NA | NA | 0 | 0.2 | NA | 1 | 0 | 0.20 | 0.00 | 0.0 | 8e-02 | 0.050 | 5e-02 |
| Foragefish1 | 0 | 5.1000 | 0.610 | 3.520 | NA | NA | 0 | 0.2 | NA | 1 | 0 | 0.00 | 0.30 | 0.0 | 1e-03 | 0.050 | 0e+00 |
| Foragefish2 | 0 | 4.7000 | 0.650 | 5.650 | NA | NA | 0 | 0.2 | NA | 1 | 0 | 0.00 | 0.08 | 0.0 | 1e-03 | 0.010 | 0e+00 |
| OtherForagefish | 0 | 5.1000 | 1.500 | 3.600 | NA | NA | 0 | 0.2 | NA | 1 | 0 | 0.00 | 0.02 | 0.0 | 1e-03 | 0.010 | 0e+00 |
| Megabenthos | 0 | NA | 0.900 | 2.984 | 0.8 | NA | 0 | 0.2 | NA | 1 | 0 | 0.00 | 0.00 | 0.1 | 0e+00 | 0.000 | 9e-02 |
| Shellfish | 0 | 7.0000 | 1.300 | NA | NA | 0.25 | 0 | 0.2 | NA | 1 | 0 | 0.00 | 0.00 | 0.5 | 0e+00 | 0.000 | 1e-02 |
| Macrobenthos | 0 | 17.4000 | 7.000 | NA | NA | 0.35 | 0 | 0.2 | NA | 1 | 0 | 0.00 | 0.00 | 0.0 | 0e+00 | 0.000 | 1e-04 |
| Zooplankton | 0 | 23.0000 | 39.000 | NA | NA | 0.25 | 0 | 0.4 | NA | 1 | 0 | 0.00 | 0.00 | 0.0 | 0e+00 | 0.000 | 0e+00 |
| Phytoplankton | 1 | 10.0000 | 240.000 | NA | NA | NA | 0 | 0.0 | NA | 1 | 0 | 0.00 | 0.00 | 0.0 | 0e+00 | 0.000 | 0e+00 |
| Detritus | 2 | NA | NA | NA | NA | NA | 0 | 0.0 | 0 | 0 | 0 | 0.00 | 0.00 | 0.0 | 0e+00 | 0.000 | 0e+00 |
| Discards | 2 | NA | NA | NA | NA | NA | 0 | 0.0 | 0 | 0 | 0 | 0.00 | 0.00 | 0.0 | 0e+00 | 0.000 | 0e+00 |
| Trawlers | 3 | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 1 | NA | NA | NA | NA | NA | NA |
| Midwater | 3 | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 1 | NA | NA | NA | NA | NA | NA |
| Dredgers | 3 | NA | NA | NA | NA | NA | NA | NA | NA | 0 | 1 | NA | NA | NA | NA | NA | NA |
Stanza Parameters
You may have noticed that the biomass and consumption to biomass parameters are missing from some of the multistanza groups. Similar to EwE, Rpath calculates those parameters to ensure that stanza groups support one another (Christensen and Walters (2004)). In order to do this, you need to populate the stanza list. As mentioned earlier, this is actually a list itself containing 3 things: the number of stanza groups, stanza group parameters, and individual stanza parameters. The number of stanzas is automatically populated. For stanza groups you need their von Bertalanffy growth function specialized K and weight at 50% maturity divided by their weight infinity (relative weight at maturity). Individual stanzas need the first and last month the species is in the stanza, the total mortality (Z) on the stanza, and whether or not it is the leading stanza.
# Group parameters
REco.params$stanzas$stgroups[, VBGF_Ksp := c(0.145, 0.295, 0.0761, 0.112)]
REco.params$stanzas$stgroups[, Wmat := c(0.0769, 0.561, 0.117, 0.321)]
# Individual stanza parameters
REco.params$stanzas$stindiv[, First := c(rep(c(0, 24), 3), 0, 48)]
REco.params$stanzas$stindiv[, Last := c(rep(c(23, 400), 3), 47, 400)]
REco.params$stanzas$stindiv[, Z := c(2.026, 0.42, 2.1, 0.425, 1.5,
0.26, 1.1, 0.18)]
REco.params$stanzas$stindiv[, Leading := rep(c(F, T), 4)]| StGroupNum | StanzaGroup | nstanzas | VBGF_Ksp | VBGF_d | Wmat | BAB | RecPower |
|---|---|---|---|---|---|---|---|
| 1 | Roundfish1 | 2 | 0.1450 | 0.66667 | 0.0769 | 0 | 1 |
| 2 | Roundfish2 | 2 | 0.2950 | 0.66667 | 0.5610 | 0 | 1 |
| 3 | Flatfish1 | 2 | 0.0761 | 0.66667 | 0.1170 | 0 | 1 |
| 4 | Flatfish2 | 2 | 0.1120 | 0.66667 | 0.3210 | 0 | 1 |
| StGroupNum | StanzaNum | GroupNum | Group | First | Last | Z | Leading |
|---|---|---|---|---|---|---|---|
| 1 | 0 | 4 | JuvRoundfish1 | 0 | 23 | 2.026 | FALSE |
| 1 | 0 | 5 | AduRoundfish1 | 24 | 400 | 0.420 | TRUE |
| 2 | 0 | 6 | JuvRoundfish2 | 0 | 23 | 2.100 | FALSE |
| 2 | 0 | 7 | AduRoundfish2 | 24 | 400 | 0.425 | TRUE |
| 3 | 0 | 8 | JuvFlatfish1 | 0 | 23 | 1.500 | FALSE |
| 3 | 0 | 9 | AduFlatfish1 | 24 | 400 | 0.260 | TRUE |
| 4 | 0 | 10 | JuvFlatfish2 | 0 | 47 | 1.100 | FALSE |
| 4 | 0 | 11 | AduFlatfish2 | 48 | 400 | 0.180 | TRUE |
The final month of the ultimate stanza can be set to any value. The
function rpath.stanzas() will calculate the final month as
the point where the species reaches 90% Winf. The function
rpath.stanzas() will also add data tables containing the
weight, number, and consumption at age for each stanza group.
REco.params <- rpath.stanzas(REco.params)Table: Completed stanzas table
| age | WageS | QageS | Survive | B | Q | NageS |
|---|---|---|---|---|---|---|
| 0 | 0.0000000 | 0.0000000 | 1.0000000 | 0.00e+00 | 0.0000000 | 23.43477 |
| 1 | 0.0000017 | 0.0001442 | 0.8446497 | 1.50e-06 | 0.0001218 | 19.79417 |
| 2 | 0.0000136 | 0.0005700 | 0.7134331 | 9.70e-06 | 0.0004067 | 16.71914 |
| 3 | 0.0000451 | 0.0012673 | 0.6026010 | 2.72e-05 | 0.0007637 | 14.12181 |
| 4 | 0.0001050 | 0.0022261 | 0.5089867 | 5.35e-05 | 0.0011330 | 11.92799 |
| 5 | 0.0002015 | 0.0034369 | 0.4299155 | 8.66e-05 | 0.0014776 | 10.07497 |
Output from the rpath.stanzas() function can be plotted
using the stanzaplot() function.
stanzaplot(REco.params, StanzaGroup = 1)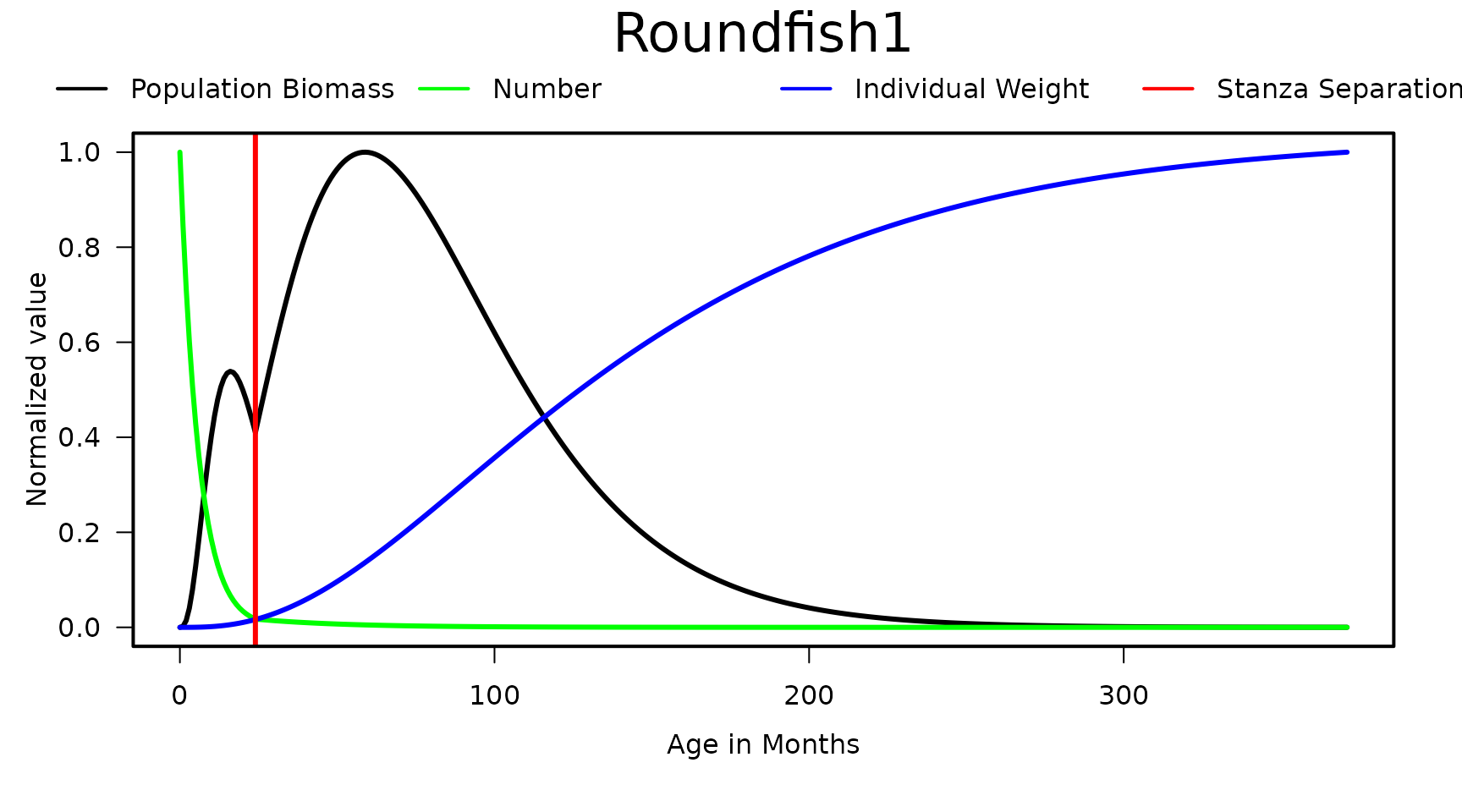
Example plot using function stanzaplot() with StanzaGroup=1
Note: If you do not have multistanza groups in your model, you
do not have to run rpath.stanzas().
Diet Parameters
The data entered in the diet list is the same as the data entered in
the diet composition tab in EwE. Just as within EwE, the columns
represent the predators while the rows represent the prey. Individual
diet components can be adjusted by specifying the prey in the
Group variable and assigning a value to the predator. For
example, if you wanted to assign 10% of the seabird diet as ‘Other
Groundfish’ you could do it like this:
REco.params$diet[Group == 'OtherGroundfish', Seabirds := 0.1]You can also assign the entire diet composition for a predator:
whale.diet <- c(rep(NA, 3), 0.01, NA, 0.01, NA, 0.01, NA, 0.01, rep(NA, 4), 0.1,
rep(NA, 3), 0.86, rep(NA, 3), NA)
REco.params$diet[, Whales := whale.diet]| Group | Seabirds | Whales |
|---|---|---|
| Seabirds | NA | NA |
| Whales | NA | NA |
| Seals | NA | NA |
| JuvRoundfish1 | NA | 0.01 |
| AduRoundfish1 | NA | NA |
| JuvRoundfish2 | NA | 0.01 |
| AduRoundfish2 | NA | NA |
| JuvFlatfish1 | NA | 0.01 |
| AduFlatfish1 | NA | NA |
| JuvFlatfish2 | NA | 0.01 |
| AduFlatfish2 | NA | NA |
| OtherGroundfish | 0.1 | NA |
| Foragefish1 | NA | NA |
| Foragefish2 | NA | NA |
| OtherForagefish | NA | 0.10 |
| Megabenthos | NA | NA |
| Shellfish | NA | NA |
| Macrobenthos | NA | NA |
| Zooplankton | NA | 0.86 |
| Phytoplankton | NA | NA |
| Detritus | NA | NA |
| Discards | NA | NA |
| Import | NA | NA |
Here is the completed model parameter file for R Ecosystem:
REco.params$diet[, Seabirds := c(rep(NA, 11), 0.1, 0.25, 0.2, 0.15,
rep(NA, 6), 0.3, NA)]
REco.params$diet[, Whales := c(rep(NA, 3), 0.01, NA, 0.01, NA, 0.01,
NA, 0.01, rep(NA, 4), 0.1, rep(NA, 3),
0.86, rep(NA, 3), NA)]
REco.params$diet[, Seals := c(rep(NA, 3), 0.05, 0.1, 0.05, 0.2, 0.005,
0.05, 0.005, 0.01, 0.24, rep(0.05, 4),
0.09, rep(NA, 5), NA)]
REco.params$diet[, JuvRoundfish1 := c(rep(NA, 3), rep(c(1e-4, NA), 4), 1e-3,
rep(NA, 2), 0.05, 1e-4, NA, .02, 0.7785,
0.1, 0.05, NA, NA)]
REco.params$diet[, AduRoundfish1 := c(rep(NA, 5), 1e-3, 0.01, 1e-3, 0.05, 1e-3,
0.01, 0.29, 0.1, 0.1, 0.347, 0.03, NA,
0.05, 0.01, rep(NA, 3), NA)]
REco.params$diet[, JuvRoundfish2 := c(rep(NA, 3), rep(c(1e-4, NA), 4), 1e-3,
rep(NA, 2), 0.05, 1e-4, NA, .02, 0.7785,
0.1, .05, NA, NA)]
REco.params$diet[, AduRoundfish2 := c(rep(NA, 3), 1e-4, NA, 1e-4, NA, rep(1e-4, 4),
0.1, rep(0.05, 3), 0.2684, 0.01, 0.37, 0.001,
NA, 0.1, NA, NA)]
REco.params$diet[, JuvFlatfish1 := c(rep(NA, 3), rep(c(1e-4, NA), 4), rep(NA, 3),
rep(1e-4, 2), NA, 0.416, 0.4334, 0.1, 0.05,
NA, NA)]
REco.params$diet[, AduFlatfish1 := c(rep(NA, 7), rep(1e-4, 5), rep(NA, 2), 0.001,
0.05, 0.001, 0.6, 0.2475, NA, 0.1, NA, NA)]
REco.params$diet[, JuvFlatfish2 := c(rep(NA, 3), rep(c(1e-4, NA), 4), rep(NA, 3),
rep(1e-4, 2), NA, 0.416, 0.4334, 0.1, 0.05,
NA, NA)]
REco.params$diet[, AduFlatfish2 := c(rep(NA, 7), 1e-4, NA, 1e-4, rep(NA, 4),
rep(1e-4, 3), 0.44, 0.3895, NA, 0.17, NA, NA)]
REco.params$diet[, OtherGroundfish := c(rep(NA, 3), rep(1e-4, 8), 0.05, 0.08, 0.0992,
0.3, 0.15, 0.01, 0.3, 0.01, rep(NA, 3), NA)]
REco.params$diet[, Foragefish1 := c(rep(NA, 3), rep(c(1e-4, NA), 4), rep(NA, 7),
0.8196, 0.06, 0.12, NA, NA)]
REco.params$diet[, Foragefish2 := c(rep(NA, 3), rep(c(1e-4, NA), 4), rep(NA, 7),
0.8196, 0.06, 0.12, NA, NA)]
REco.params$diet[, OtherForagefish := c(rep(NA, 3), rep(c(1e-4, NA), 4), rep(NA, 7),
0.8196, 0.06, 0.12, NA, NA)]
REco.params$diet[, Megabenthos := c(rep(NA, 15), 0.1, 0.03, 0.55, rep(NA, 2), 0.32,
NA, NA)]
REco.params$diet[, Shellfish := c(rep(NA, 18), 0.3, 0.5, 0.2, NA, NA)]
REco.params$diet[, Macrobenthos := c(rep(NA, 16), 0.01, rep(0.2, 2), NA, 0.59, NA, NA)]
REco.params$diet[, Zooplankton := c(rep(NA, 18), 0.2, 0.6, 0.2, NA, NA)]| Group | Seabirds | Whales | Seals | JuvRoundfish1 | AduRoundfish1 | JuvRoundfish2 | AduRoundfish2 | JuvFlatfish1 | AduFlatfish1 | JuvFlatfish2 | AduFlatfish2 | OtherGroundfish | Foragefish1 | Foragefish2 | OtherForagefish | Megabenthos | Shellfish | Macrobenthos | Zooplankton | Phytoplankton |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Seabirds | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Whales | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Seals | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| JuvRoundfish1 | NA | 0.01 | 0.050 | 0.0001 | NA | 0.0001 | 0.0001 | 0.0001 | NA | 0.0001 | NA | 0.0001 | 0.0001 | 0.0001 | 0.0001 | NA | NA | NA | NA | NA |
| AduRoundfish1 | NA | NA | 0.100 | NA | NA | NA | NA | NA | NA | NA | NA | 0.0001 | NA | NA | NA | NA | NA | NA | NA | NA |
| JuvRoundfish2 | NA | 0.01 | 0.050 | 0.0001 | 0.001 | 0.0001 | 0.0001 | 0.0001 | NA | 0.0001 | NA | 0.0001 | 0.0001 | 0.0001 | 0.0001 | NA | NA | NA | NA | NA |
| AduRoundfish2 | NA | NA | 0.200 | NA | 0.010 | NA | NA | NA | NA | NA | NA | 0.0001 | NA | NA | NA | NA | NA | NA | NA | NA |
| JuvFlatfish1 | NA | 0.01 | 0.005 | 0.0001 | 0.001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | NA | NA | NA | NA | NA |
| AduFlatfish1 | NA | NA | 0.050 | NA | 0.050 | NA | 0.0001 | NA | 0.0001 | NA | NA | 0.0001 | NA | NA | NA | NA | NA | NA | NA | NA |
| JuvFlatfish2 | NA | 0.01 | 0.005 | 0.0001 | 0.001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | 0.0001 | NA | NA | NA | NA | NA |
| AduFlatfish2 | NA | NA | 0.010 | NA | 0.010 | NA | 0.0001 | NA | 0.0001 | NA | NA | 0.0001 | NA | NA | NA | NA | NA | NA | NA | NA |
| OtherGroundfish | 0.10 | NA | 0.240 | 0.0010 | 0.290 | 0.0010 | 0.1000 | NA | 0.0001 | NA | NA | 0.0500 | NA | NA | NA | NA | NA | NA | NA | NA |
| Foragefish1 | 0.25 | NA | 0.050 | NA | 0.100 | NA | 0.0500 | NA | NA | NA | NA | 0.0800 | NA | NA | NA | NA | NA | NA | NA | NA |
| Foragefish2 | 0.20 | NA | 0.050 | NA | 0.100 | NA | 0.0500 | NA | NA | NA | NA | 0.0992 | NA | NA | NA | NA | NA | NA | NA | NA |
| OtherForagefish | 0.15 | 0.10 | 0.050 | 0.0500 | 0.347 | 0.0500 | 0.0500 | 0.0001 | 0.0010 | 0.0001 | 0.0001 | 0.3000 | NA | NA | NA | NA | NA | NA | NA | NA |
| Megabenthos | NA | NA | 0.050 | 0.0001 | 0.030 | 0.0001 | 0.2684 | 0.0001 | 0.0500 | 0.0001 | 0.0001 | 0.1500 | NA | NA | NA | 0.10 | NA | NA | NA | NA |
| Shellfish | NA | NA | 0.090 | NA | NA | NA | 0.0100 | NA | 0.0010 | NA | 0.0001 | 0.0100 | NA | NA | NA | 0.03 | NA | 0.01 | NA | NA |
| Macrobenthos | NA | NA | NA | 0.0200 | 0.050 | 0.0200 | 0.3700 | 0.4160 | 0.6000 | 0.4160 | 0.4400 | 0.3000 | NA | NA | NA | 0.55 | NA | 0.20 | NA | NA |
| Zooplankton | NA | 0.86 | NA | 0.7785 | 0.010 | 0.7785 | 0.0010 | 0.4334 | 0.2475 | 0.4334 | 0.3895 | 0.0100 | 0.8196 | 0.8196 | 0.8196 | NA | 0.3 | 0.20 | 0.2 | NA |
| Phytoplankton | NA | NA | NA | 0.1000 | NA | 0.1000 | NA | 0.1000 | NA | 0.1000 | NA | NA | 0.0600 | 0.0600 | 0.0600 | NA | 0.5 | NA | 0.6 | NA |
| Detritus | NA | NA | NA | 0.0500 | NA | 0.0500 | 0.1000 | 0.0500 | 0.1000 | 0.0500 | 0.1700 | NA | 0.1200 | 0.1200 | 0.1200 | 0.32 | 0.2 | 0.59 | 0.2 | NA |
| Discards | 0.30 | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| Import | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
Pedigree parameters
Rpath does not currently use pedigrees. However, future Rpath extensions will use them. Therefore, we include them in the current parameter object. The default values are 1 (low confidence). These defaults are not changed for R Ecosystem but can obviously be changed in a similar manner to the other parameter files.
| Group | Biomass | PB | QB | Diet | Trawlers | Midwater | Dredgers |
|---|---|---|---|---|---|---|---|
| Seabirds | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Whales | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Seals | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| JuvRoundfish1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| AduRoundfish1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| JuvRoundfish2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| AduRoundfish2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| JuvFlatfish1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| AduFlatfish1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| JuvFlatfish2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| AduFlatfish2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| OtherGroundfish | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Foragefish1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Foragefish2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| OtherForagefish | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Megabenthos | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Shellfish | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Macrobenthos | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Zooplankton | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Phytoplankton | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Detritus | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Discards | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Trawlers | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Midwater | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Dredgers | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
